Documentation aside, things move forward
Yesterday was the first day of testing.
Today, I fixed 2 things I considered key.
Overwriting the defaults for prints and plots
using the C++ component for matching (and thereby fixing a bit of parallel processing!)
Both I describe next.
Easy or not so easy: S3Methods overwrites
I will say: It gets confusing to me, whether/how/when Roxyen2 will be key… And when not?
I know the thing works now with a mix of roxygen2
#' @export
print.rlcs_rule <- function(x, ...) {
print(paste(x$condition_string, x$action), ...)
}But also required was an EDIT to NAMESPACE (which roxygen somehow then complains about?)
S3method(print,rlcs_rule)
S3method(print,rlcs_population)
S3method(plot,rlcs_population)Both together, at least, do work as expected.
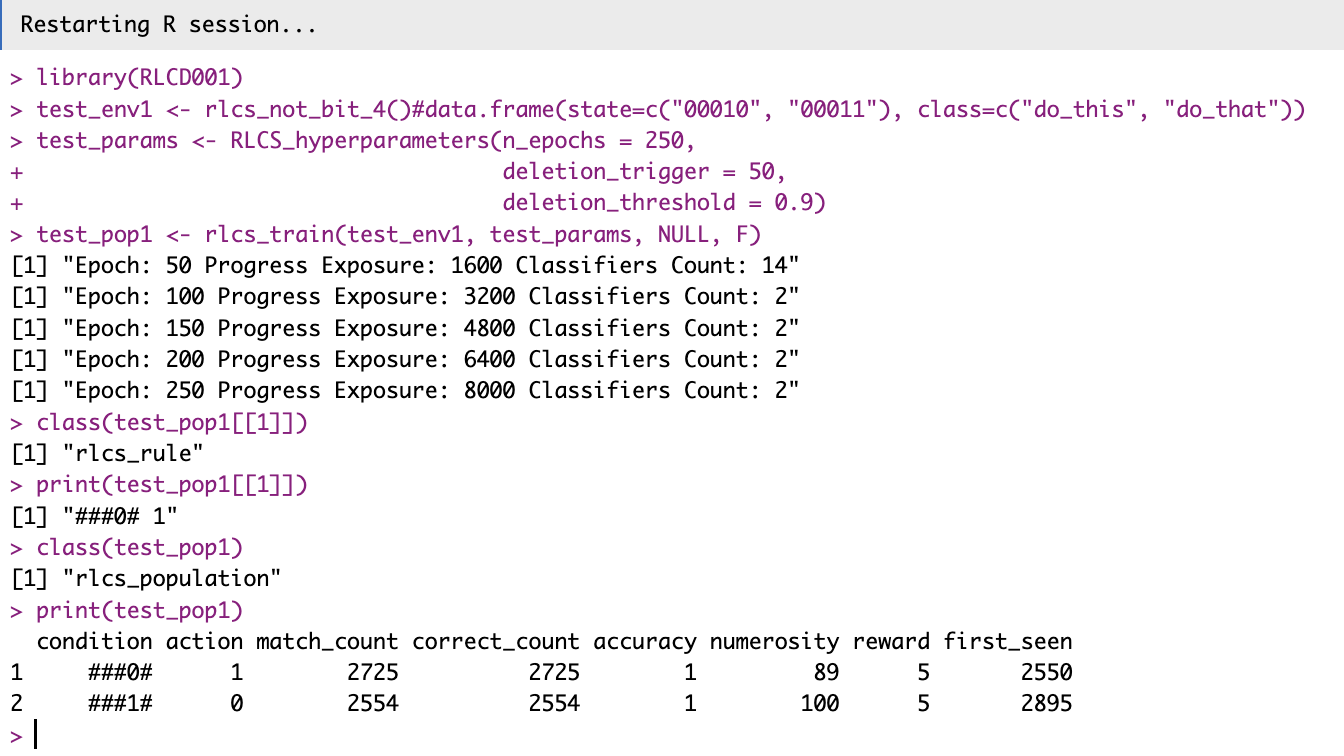
Using C++ was not difficult
Just… Put your correctly formatted .cpp file in the src/ directory, and RStudio will know how to go about it (supposing you created your RStudio project with C++ support, at least).
And I know it works, because I made sure the function for matching uses it, and more importantly…
It fixed something else
Parallelizing with %dopar% with the C++ version failed before, but with the package loaded, it now works seamlessly!

Conclusion
My next focus should be on clean-up and documentation. And maybe a better understanding of the Roxygen2 role and NAMESPACE (for exporting only certain functions, I believe…).
But overall, I’m back on track with the RLCS package :)